Introduction
Searching Large Language Model (LLM) databases for information can take quite a long time if the database is large. Restricting the search to a relevant subset can sped up response time, but of course with a risk that relevant information will not be looked at.
AllegroGraph provides two methods to restirct the search, and if well chosen, these can speed up responses with little or no loss of information.
The two methods are to use a ?selector (see ?selector introduction) and the second is to ?useClustering (see ?useClustering).
?selector introduction
Suppose you want to build a vector database of medical terminology categorized as follows:
- Conditions
- Observations
- Medications
- Procedures
When the query is "aspirin tablet" it may make sense to search only the Medications category, or when "Heart Attack" only the Conditions category. If the four categories are about equally populated, and the you can specify that just one category be searched, the vector matching process will run 4 times faster. In AllegroGraph, the selector argument enables this efficiency.
AllegroGraph provides three Magic predicates that access a vector repository: llm:nearestNeighbor, llm:askMyDocuments and llm:chatState.
These predicates have a special argument, ?selector, that enables searching only a subset of the vector repo, rather than all the embeddings. Depending on the number of embeddings found by a SPARQL query, vector matching speed may increase dramatically.
Selector Form
The selector is a string containing the body of a SPARQL query that binds the variable ?id to a subset of the vector database objects. These selections are typically based on object properties, a feature of the vector repository.
For example, to select only the vectors associated with the category Medication, we can write
PREFIX vdbp: <http://franz.com/vdb/prop/>
PREFIX health: <http://local.com/health/>
SELECT * {
(?vid ?score ?text)
llm:nearestNeighbor("A non-aspirin pain medication" "vectorRepo"
:selector "{?id vdbp:category health:Medication}")
} Note that the predicate binds the output value ?vid to the subset of ?ids returned by the nearest-neighbor operation. Additionally, the selector may use keyword syntax, identified by the key :selector
As stated above, the value of the selector is a string containing the body of a valid SPARQL query that binds the variable ?id. That SPARQL query may be as complex as necessary. For example a valid selector could be:
:selector
"{?id prop:category category:Product.
?id prop:quantity ?count.
?id prop:price ?price.
FILTER (?count > 100 && ?price < 10)
}" As this example implies, the selector SPARQL may utilize any namespaces declared in the query environment:
:selector
"{?id prop:kingdom example:Animal}" The body of the SPARQL query should begin and end with curly braces {...}.
No out-of-body operations such as DELETE, INSERT or ORDER BY are permitted. SELECT need not be specified because the selector query implicitly wraps the body with
SELECT ?id {...} Examples
The following examples illustrate the selector using an OpenAI model.
Example of selector with nearestNeighbor
This example demonstrates the use of a selector to choose terms naming either animals or plants.
We'll generate some synthetic data to demonstrate the use of the selector argument. In a real-world scenario, you would replace the sample data with data from your company databases and/or private and public datasets.
To begin, create a repo named example. You can choose whatever name you wish, but we'll use example.
Use the llm:response predicate to generate the sample data:
PREFIX franzOption_openaiApiKey: <franz:OPENAI_API_KEY_HERE>
PREFIX val: <http://local.com/value/>
DELETE {?s ?p ?o} WHERE {?s ?p ?o};
INSERT {?anode rdf:type val:Animal.
?anode skos:prefLabel ?animal.
} WHERE {
?animal llm:response "List 256 common animals."
bind(llm:node(?animal) as ?anode).};
INSERT {?anode rdf:type val:Plant.
?anode skos:prefLabel ?plant.
} WHERE {
?plant llm:response "List 256 common plants."
bind(llm:node(?plant) as ?anode).}; Note that the query performs a DELETE operation in order to ensure the repository is empty.
One way to generate the embeddings for a vector repo is the Webview embedding tool. To find the tool, navigate to Repository --> Repository Control --> Create LLM Embeddings
Another method uses the agtool command with the llm index arguments, as we show below.
We'll illustrate the Webview approach in the next example. Either method provides for the insertion of property values alongside each embedded object.
To utilize agtool, first create two vector repo definition files animal.def and plant.def:
The file animal.def contains
gpt
embedder openai
if-exists supersede
api-key "OPENAI_API_KEY_HERE"
vector-database-name exampleVecs
include-predicates <http://www.w3.org/2004/02/skos/core\#prefLabel>
include-types <http://local.com/value/Animal>
property kingdom Animal And plant.def contains
gpt
embedder openai
if-exists open
api-key "OPENAI_API_KEY_HERE"
vector-database-name exampleVecs
include-predicates <http://www.w3.org/2004/02/skos/core\#prefLabel>
include-types <http://local.com/value/Plant>
property kingdom Plant After creating the definition files, run
agtool llm index example animal.def and
agtool llm index example plant.def Notice that the first (animal) definition file causes agtool to create or supersede a vector store (because it contains if-exists supersede) while the second (plant) definition file merely opens it (as it contains if-exists open).
Now we are ready to test nearestNeighbor on the vector repository.
PREFIX franzOption_openaiApiKey: <franz:OPENAI_API_KEY_HERE>
PREFIX vdbp: <http://franz.com/vdb/prop/>
SELECT DISTINCT ?text ?score {
(?id ?score ?text)
llm:nearestNeighbor
("They are beautiful" "exampleVecs"
:minScore 0.0 :topN 5 ) } The top 5 results are:
Magnolia
Iris
Bird of Paradise
Swan
Peacock (Note: Your results may differ because the LLM synthetic data varies randomly)
We can specify the selector so only plants or only animals are considered, for example this query looks for beautiful plants:
With the query
PREFIX franzOption_openaiApiKey: <franz:OPENAI_API_KEY_HERE>
PREFIX vdbp: <http://franz.com/vdb/prop/>
SELECT ?text {
(?id ?score ?text)
llm:nearestNeighbor
("They are beautiful"
"exampleVecs"
:minScore 0.0 :topN 5
:selector
"""{
?id vdbp:kingdom "Plant".
}""") } The top 5 results are
Magnolia
Iris
Bird of Paradise
Primrose
Violet The selector allows the query to search only the objects in one category or the other. The selector query runs faster because the llm:nearestNeighbor predicate has to search fewer objects.
In a real-world use case, there could be many more properties (e.g. Condition, Observation, Medication and Procedure in a repository of medical terminology). The selector may include any or all of these properties.
Example selector with askMyDocuments
The :selector keyword argument in llm:askMyDocuments selects RAG documents from a subset of a vector repo, based on property values. In this example we create a vector repo containing true facts and fantasies, and use the world property to select one or the other: Reality or Fantasy.
First, generate some sample document data using an LLM prompt.
Create a repo called myDocuments. Run this query to insert some document data.
PREFIX franzOption_openaiApiKey: <franz:YOUR_OPENAI_API_KEY_HERE>
PREFIX val: <http://local.com/value/>
DELETE {?s ?p ?o} WHERE
{?s ?p ?o};
INSERT {?fnode rdf:type val:Fantasy.
?fnode skos:prefLabel ?fantasy.
?rnode rdf:type val:Reality.
?rnode skos:prefLabel ?reality.
}
WHERE {
(?fantasy ?reality) llm:askForTable "List 40 facts true in the Harry Potter world but untrue in the real world, and the real truth about those facts".
bind(llm:node(?fantasy) as ?fnode).
bind(llm:node(?reality) as ?rnode).
} Next, create embeddings for the vector repository.
In Webview, Navigate to Repository --> RepositoryControl --> Create LLM Embedding.
Create embeddings in two steps: one for Fantasy, and one for Reality. The yellow highlights indicate the differences between the two embedding runs.
Fantasy
Fill in the form on the page Create LLM Embeddings with these values:
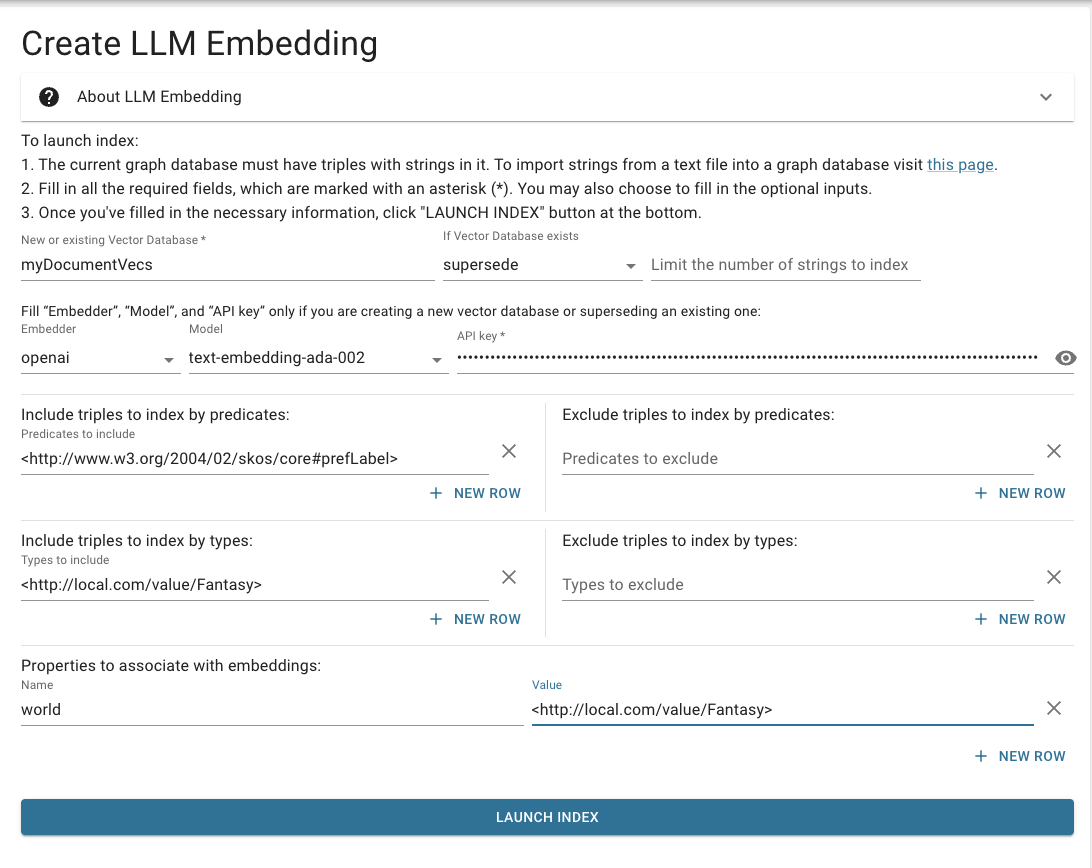
Click: Launch Index
Wait for indexing to finish.
Reality
When embedding finishes, replace the form fields with:
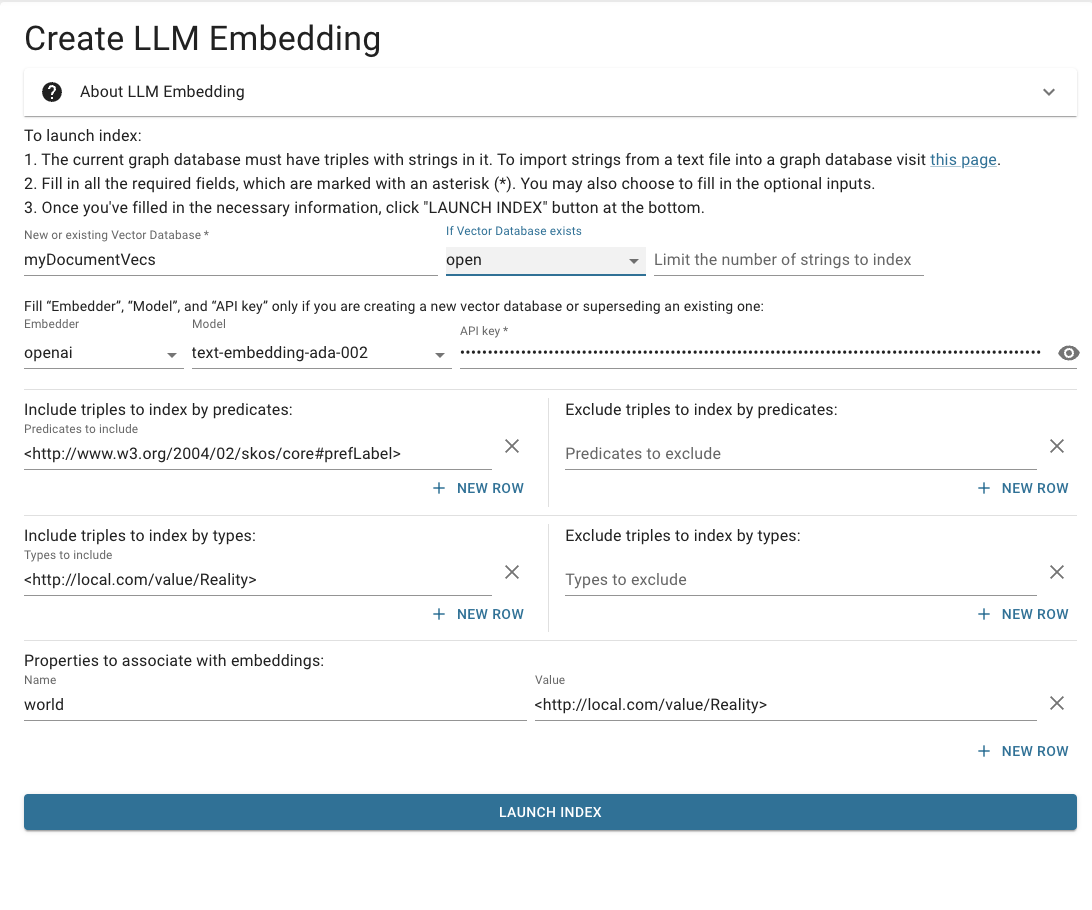
It's very important to notice that the Fantasy embeddings supersede an existing database, and the Reality embeddings only open it.
Click: Launch Index.
After the embedding finishes, you can query using the selector:
PREFIX franzOption_openaiApiKey: <franz:YOUR_OPENAI_API_KEY_HERE>
SELECT * {
(?response ?score ?citationId ?originalText)
llm:askMyDocuments
("I want to travel back in time" "myDocumentVecs" :minScore 0.0
:selector
"{?id <http://franz.com/vdb/prop/world> <http://local.com/value/Reality>}"
).
} The :selector argument is optional. Depending on the choice of selector, you receive different responses:
- No selector:
"Time-turners are fictional objects from the Harry Potter world, allowing witches and wizards to travel back in time. In the real world, time travel is not possible as magic does not exist."
- Select Fantasy:
:selector "{?id <http://franz.com/vdb/prop/world> <http://local.com/value/Fantasy>}" - Select Reality:
:selector "{?id <http://franz.com/vdb/prop/world> <http://local.com/value/Reality>}"
"Time-turners in the magical world allow witches and wizards to travel back in time."
"Time-travel is a fictional concept, and in reality, it is not possible to travel back in time."
?useClustering introduction
Vector databases can be divided in clusterings, that is partitioned into disjoint subsets. These can then be used with certain queries using LLM magic properties (llm:nearestNeighbor, llm:askMyDocuments and llm:chatState) to speed up the response. Clusterings will be used is the ?useClustering optional input argument is specified true.
Using agtool to create clusters is described in the section Clustering objects for search using agtool and the ?useClustering argument of the Embedding Knowledge in a Vector Database document.